text_mining
This is an old revision of the document!
Table of Contents
E.g. 2 mode matrix data
Load data
setwd("d:/rdata")
load("data/termDocMatrix.rdata") # load termDocMatrix
termDocMatrix[5:10,1:20] # inspect part of the matrix
> load("termDocMatrix.rdata") # load termDocMatrix
> termDocMatrix[5:10,1:20] # inspect part of the matrix
Docs
Terms 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20
data 1 1 0 0 1 0 0 0 0 0 1 1 1 1 1 0 1 0 0 0
examples 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
introduction 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1
mining 0 0 0 0 0 0 0 0 0 0 0 1 1 0 1 0 0 0 0 0
network 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 1 1 1
package 0 0 0 1 1 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0
Terms x Documents matrix data = two mode matrix data
Transform Data into an Adjacency Matrix
termDocMatrix[termDocMatrix>=1] <- 1 # change it to a Boolean matrix termMatrix <- termDocMatrix %*% t(termDocMatrix) # transform into a term-term adjacency matrix termMatrix[5:10,5:10] # inspect terms numbered 5 to 10
> termDocMatrix[termDocMatrix>=1] <- 1 # change it to a Boolean matrix
> termMatrix <- termDocMatrix %*% t(termDocMatrix) # transform into a term-term adjacency matrix
> termMatrix[5:10,5:10] # inspect terms numbered 5 to 10
Terms
Terms data examples introduction mining network package
data 53 5 2 34 0 7
examples 5 17 2 5 2 2
introduction 2 2 10 2 2 0
mining 34 5 2 47 1 5
network 0 2 2 1 17 1
package 7 2 0 5 1 21
>
Two mode → one mode data by termMatrix = termDocmatrix * transposed(termDocmatrix)
- termMatrix data = one mode matrix data showing the relationships among the words (appeared in the Doc)
- For example, the word, “data” appears in the doc a toal of “53”
- And, In a total of “5” cases, both data and examples appeared at the same time(document).
- Therefore, the word, “mining(34)” is more close (relevant) to “data” than “examples(5)”
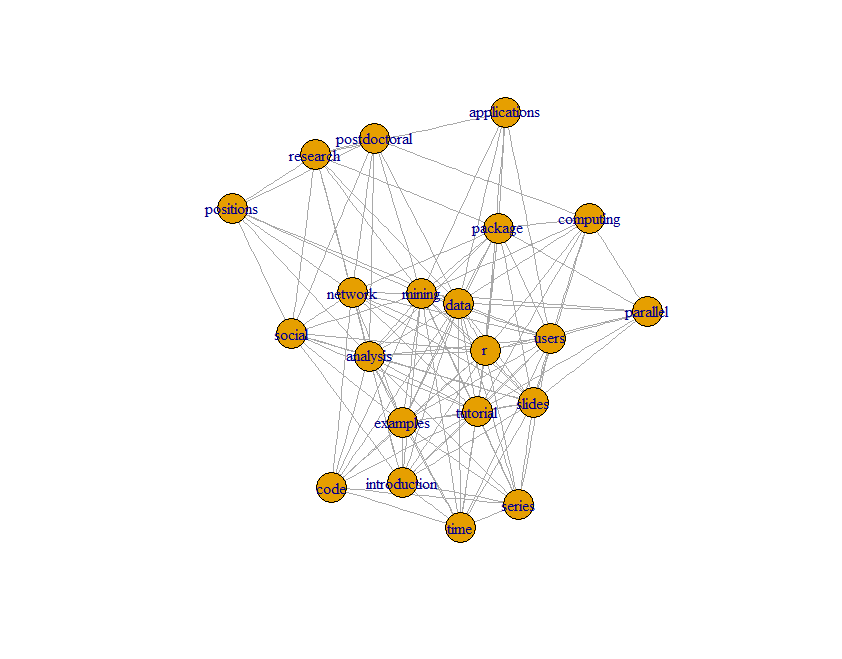
Next we can build a graph with graph.adjacency() from package igraph.
library(igraph) # build a graph from the above matrix g <- graph.adjacency(termMatrix, weighted=T, mode = “undirected”) # remove loops g <- simplify(g) # set labels and degrees of vertices V(g)$label <- V(g)$name V(g)$degree <- degree(g) V(g)$label V(g)$degree
> library(igraph) > g <- graph.adjacency(termMatrix, weighted=T, mode = "undirected") > g <- simplify(g) > V(g)$label <- V(g)$name > V(g)$degree <- degree(g) > V(g)$degree [1] 17 6 9 9 18 14 12 20 14 13 8 7 8 17 9 11 15 11 11 16 15 > V(g)$label [1] "analysis" "applications" "code" "computing" [5] "data" "examples" "introduction" "mining" [9] "network" "package" "parallel" "positions" [13] "postdoctoral" "r" "research" "series" [17] "slides" "social" "time" "tutorial" [21] "users"
Plot a Graph
# set seed to make the layout reproducible set.seed(3952) layout1 <- layout.fruchterman.reingold(g) plot(g, layout=layout1)
> # set seed to make the layout reproducible > set.seed(3952) > layout1 <- layout.fruchterman.reingold(g) > plot(g, layout=layout1)
Different layout of plot
plot(g, layout=layout.kamada.kawai) tkplot(g, layout=layout.kamada.kawai)
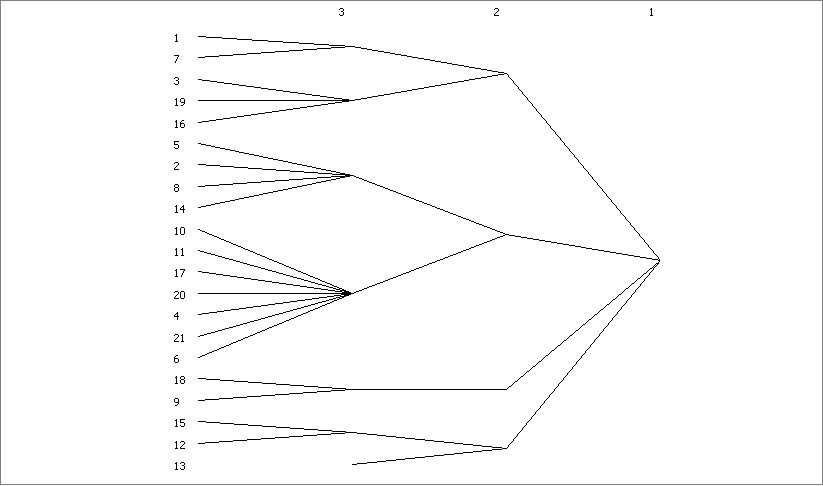
CONCOR
--------------------------------------------------------------------------------
Diagonal: Ignore
Max partitions: 3
Input dataset: terms (D:\Users\Hyo\Documents\UCINET data\rdm\terms)
Initial Correlation Matrix
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21
analy appli code compu data examp intro minin netwo packa paral posit postd r resea serie slide socia time tutor users
----- ----- ----- ----- ----- ----- ----- ----- ----- ----- ----- ----- ----- ----- ----- ----- ----- ----- ----- ----- -----
1 analysis 1.00 0.19 0.49 0.22 0.18 0.51 0.73 0.23 0.75 0.43 0.13 0.13 0.10 0.07 0.02 0.26 0.59 0.71 0.26 0.73 0.47
2 applications 0.19 1.00 0.36 0.28 0.97 0.56 0.38 0.98 -0.14 0.68 0.26 0.38 0.50 0.72 0.50 0.17 0.63 -0.16 0.17 0.55 0.51
3 code 0.49 0.36 1.00 0.45 0.53 0.92 0.47 0.38 0.13 0.67 0.35 -0.22 -0.17 0.40 -0.22 0.55 0.67 0.05 0.55 0.62 0.75
4 computing 0.22 0.28 0.45 1.00 0.35 0.56 0.03 0.32 0.03 0.76 0.97 -0.22 -0.27 0.20 -0.19 0.16 0.61 -0.11 0.16 0.44 0.71
5 data 0.18 0.97 0.53 0.35 1.00 0.56 0.28 0.94 -0.00 0.64 0.30 -0.00 0.29 0.54 0.18 0.28 0.59 -0.06 0.28 0.49 0.60
6 examples 0.51 0.56 0.92 0.56 0.56 1.00 0.47 0.60 0.26 0.83 0.47 -0.11 -0.07 0.68 -0.10 0.54 0.85 0.11 0.54 0.76 0.84
7 introduction 0.73 0.38 0.47 0.03 0.28 0.47 1.00 0.37 0.48 0.42 -0.01 0.07 0.17 0.52 -0.01 0.41 0.67 0.36 0.41 0.68 0.48
8 mining 0.23 0.98 0.38 0.32 0.94 0.60 0.37 1.00 -0.05 0.71 0.28 0.41 0.31 0.67 0.51 0.19 0.62 -0.13 0.19 0.52 0.54
9 network 0.75 -0.14 0.13 0.03 -0.00 0.26 0.48 -0.05 1.00 0.14 -0.03 0.13 0.21 -0.09 -0.01 0.15 0.24 0.92 0.15 0.39 0.25
10 package 0.43 0.68 0.67 0.76 0.64 0.83 0.42 0.71 0.14 1.00 0.68 -0.01 0.02 0.84 0.03 0.32 0.88 -0.02 0.32 0.77 0.91
11 parallel 0.13 0.26 0.35 0.97 0.30 0.47 -0.01 0.28 -0.03 0.68 1.00 -0.28 -0.21 0.32 -0.23 0.10 0.54 -0.16 0.10 0.43 0.65
12 positions 0.13 0.38 -0.22 -0.22 -0.00 -0.11 0.07 0.41 0.13 -0.01 -0.28 1.00 0.90 -0.00 0.94 -0.26 -0.06 0.30 -0.26 0.01 -0.21
13 postdoctoral 0.10 0.50 -0.17 -0.27 0.29 -0.07 0.17 0.31 0.21 0.02 -0.21 0.90 1.00 0.15 0.87 -0.19 0.01 0.28 -0.19 0.09 -0.15
14 r 0.07 0.72 0.40 0.20 0.54 0.68 0.52 0.67 -0.09 0.84 0.32 -0.00 0.15 1.00 0.13 0.22 0.72 -0.16 0.22 0.74 0.80
15 research 0.02 0.50 -0.22 -0.19 0.18 -0.10 -0.01 0.51 -0.01 0.03 -0.23 0.94 0.87 0.13 1.00 -0.30 -0.06 0.05 -0.30 -0.01 -0.18
16 series 0.26 0.17 0.55 0.16 0.28 0.54 0.41 0.19 0.15 0.32 0.10 -0.26 -0.19 0.22 -0.30 1.00 0.55 0.01 1.00 0.33 0.49
17 slides 0.59 0.63 0.67 0.61 0.59 0.85 0.67 0.62 0.24 0.88 0.54 -0.06 0.01 0.72 -0.06 0.55 1.00 0.14 0.55 0.81 0.92
18 social 0.71 -0.16 0.05 -0.11 -0.06 0.11 0.36 -0.13 0.92 -0.02 -0.16 0.30 0.28 -0.16 0.05 0.01 0.14 1.00 0.01 0.41 0.12
19 time 0.26 0.17 0.55 0.16 0.28 0.54 0.41 0.19 0.15 0.32 0.10 -0.26 -0.19 0.22 -0.30 1.00 0.55 0.01 1.00 0.33 0.49
20 tutorial 0.73 0.55 0.62 0.44 0.49 0.76 0.68 0.52 0.39 0.77 0.43 0.01 0.09 0.74 -0.01 0.33 0.81 0.41 0.33 1.00 0.80
21 users 0.47 0.51 0.75 0.71 0.60 0.84 0.48 0.54 0.25 0.91 0.65 -0.21 -0.15 0.80 -0.18 0.49 0.92 0.12 0.49 0.80 1.00
PARTITION DIAGRAM
i a p
n p o
t p c p s
a r l p t o e r o t
n o i p a u m x n e s d
a d s c m a r s t p a s e s i o
l u e a i c a l o u u m o t e t c
y c c t r d t n k l i r t s p c w a i t
s t o i i a i i a l d i i e l i o r o o
i i d m e t o n g e e a n r e a r c n r
s o e e s a n g r e l s l g s s l k h s a
1 1 1 1 1 1 2 2 1 1 1 1
Level 1 7 3 9 6 5 2 8 4 0 1 7 0 4 1 6 8 9 5 2 3
----- - - - - - - - - - - - - - - - - - - - - -
3 XXX XXXXX XXXXXXX XXXXXXXXXXXXX XXX XXX .
2 XXXXXXXXX XXXXXXXXXXXXXXXXXXXXX XXX XXXXX
1 XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX XXXXXXXXX
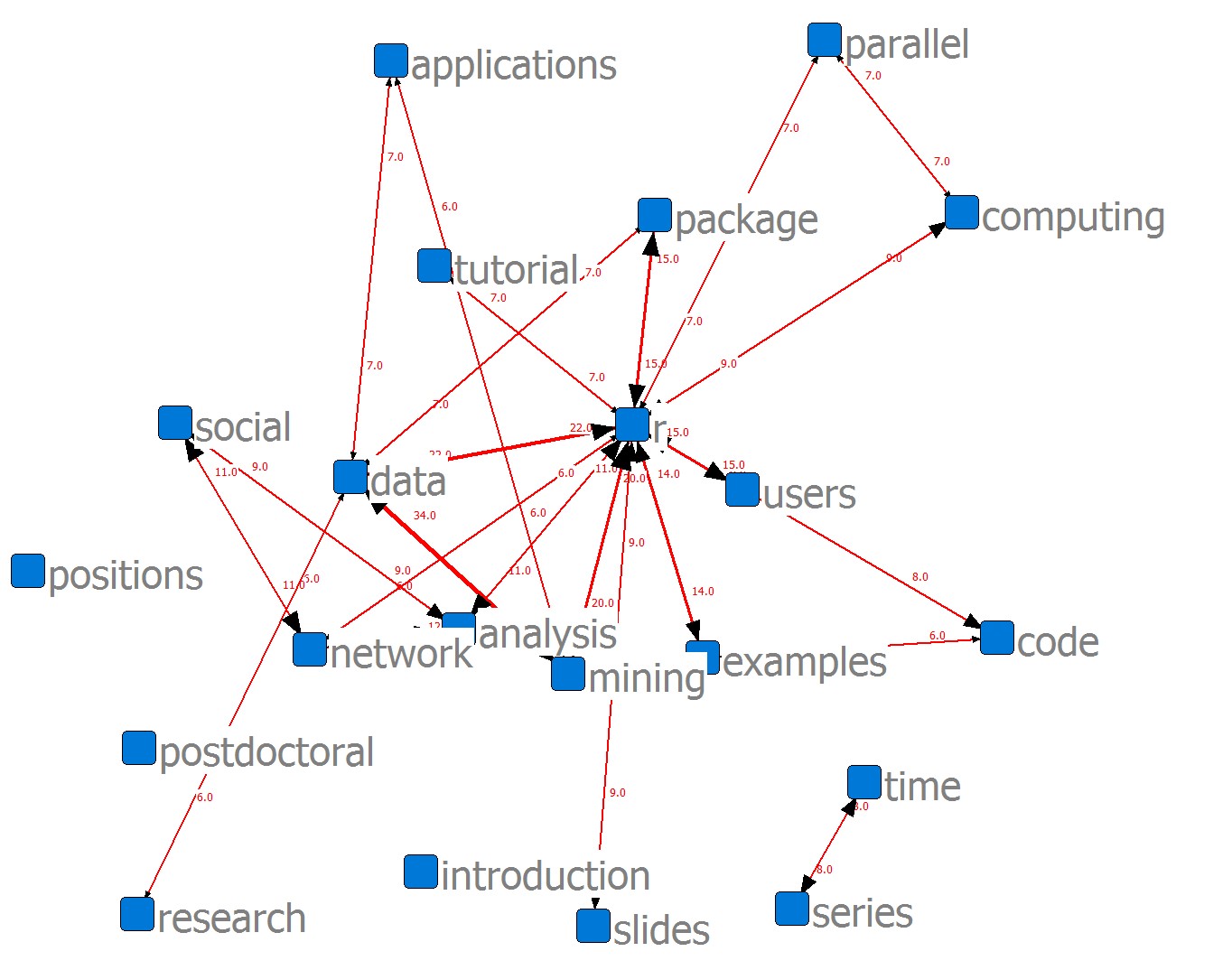
Relation Sheet 1
Blocked Matrix
1 7 3 19 16 5 2 8 14 10 11 17 20 4 21 6 18 9 15 12 13
an in co ti se da ap mi r pa pa sl tu co us ex so ne re po po
----------------------------------------------------------------------------
1 analysis | 23 2 | 1 4 4 | 4 4 11 | 2 3 4 5 4 | 9 12 | 1 2 | 3 |
7 introduction | 2 10 | 1 1 | 2 2 2 | 2 1 2 2 | 2 2 | | |
------------------------------------------------------------------------------
3 code | 1 | 9 2 2 | 1 3 8 | 1 6 | 1 | | |
19 time | 4 1 | 2 8 8 | 1 3 5 | 2 1 2 2 | | | |
16 series | 4 1 | 2 8 8 | 1 3 5 | 2 1 2 2 | | | |
------------------------------------------------------------------------------
5 data | 4 2 | 1 1 1 | 53 7 34 22 | 7 1 4 4 1 4 5 | | 6 5 | 5 |
2 applications | | | 7 9 6 4 | 1 1 | | 1 | |
8 mining | 4 2 | 3 3 3 | 34 6 47 20 | 5 1 4 4 1 5 5 | 1 1 | 2 1 | 4 |
14 r | 11 2 | 8 5 5 | 22 4 20 70 | 15 7 9 7 9 15 14 | 3 6 | | |
------------------------------------------------------------------------------
10 package | 2 | | 7 1 5 15 | 21 3 1 4 2 5 2 | 1 | 1 | |
11 parallel | | | 1 1 7 | 3 8 1 1 7 2 | | | |
17 slides | 3 2 | 2 2 | 4 4 9 | 1 1 16 1 1 4 1 | 1 2 | | |
20 tutorial | 4 1 | 1 1 1 | 4 4 7 | 4 1 1 16 1 3 3 | 2 5 | | |
4 computing | | | 1 1 9 | 2 7 1 1 10 2 | | | 1 |
21 users | 5 2 | 2 2 | 4 1 5 15 | 5 2 4 3 2 18 3 | 2 | | |
6 examples | 4 2 | 6 2 2 | 5 5 14 | 2 1 3 3 17 | 1 2 | | |
------------------------------------------------------------------------------
18 social | 9 2 | | 1 3 | 1 2 1 | 12 11 | 2 2 | 3 |
9 network | 12 2 | 1 | 1 6 | 1 2 5 2 2 | 11 17 | 1 2 | 2 |
------------------------------------------------------------------------------
15 research | 1 | | 6 1 2 | 1 | 2 1 | 12 4 | 4 |
12 positions | 2 | | 5 1 | | 2 2 | 4 11 | 4 |
------------------------------------------------------------------------------
13 postdoctoral | 3 | | 5 4 | 1 | 3 2 | 4 4 | 11 |
-----------------------------------------------------------------------------
Density Matrix
1 2 3 4 5 6 7
------ ------ ------ ------ ------ ------ ------
1 2.000 1.833 3.125 1.786 6.250 0.750 1.500
2 1.833 4.000 2.500 1.000 0.167 0.000 0.000
3 3.125 2.500 15.500 4.607 1.375 1.875 2.250
4 1.786 1.000 4.607 2.238 1.143 0.071 0.143
5 6.250 0.167 1.375 1.143 11.000 1.750 2.500
6 0.750 0.000 1.875 0.071 1.750 4.000 4.000
7 1.500 0.000 2.250 0.143 2.500 4.000
R-squared = 0.474
First order actor-by-actor correlation matrix saved as dataset Concor1stCorr
Partition-by-actor indicator matrix saved as dataset ConcorCCPart
Permutation vector saved as dataset ConcorCCPerm
----------------------------------------
Running time: 00:00:01
Output generated: 08 12 16 09:32:55
UCINET 6.614 Copyright (c) 1992-2016 Analytic Technologies
E.g. Dan McFarland's students data
# Load the 'igraph' library
library('igraph')
# (1) Read in the data files, NA data objects coded as 'na'
magact96 = read.delim('http://dl.dropbox.com/u/25710348/snaimages/mag_act96.txt', na.strings = 'na')
magact97 = read.delim('http://dl.dropbox.com/u/25710348/snaimages/mag_act97.txt', na.strings = 'na')
magact98 = read.delim('http://dl.dropbox.com/u/25710348/snaimages/mag_act98.txt', na.strings = 'na')
Variables:
- ID, gender(GND), grade(GRD), race(RCE)
- Clubs attended by the ID (1 if so, 0 if not so): Asian.Club, Hispanic.Club, . . . .
magattrib = magact96[,1:4] g96 <- as.matrix(magact96[,-(1:4)]); row.names(g96) = magact96$ID. g97 <- as.matrix(magact97[,-(1:4)]); row.names(g97) = magact97$ID. g98 <- as.matrix(magact98[,-(1:4)]); row.names(g98) = magact98$ID.
i96 <- graph.incidence(g96, mode=c('all') )
i97 <- graph.incidence(g97, mode=c('all') )
i98 <- graph.incidence(g98, mode=c('all') )
V(i96)$color[1:1295] <- rgb(1,0,0,.5) V(i96)$color[1296:1386] <- rgb(0,1,0,.5)
V(i96)$label <- V(i96)$name V(i96)$label.color <- rgb(0,0,.2,.5) V(i96)$label.cex <- .4 V(i96)$size <- 6 V(i96)$frame.color <- NA
E(i96)$color <- rgb(.5,.5,0,.2)
pdf('i96.pdf')
plot(i96, layout=layout.fruchterman.reingold)
dev.off()
i96 <- delete.vertices(i96, V(i96)[ degree(i96)==0 ])
V(i96)$label[1:857] <- NA
V(i96)$color[1:857] <- rgb(1,0,0,.1)
V(i96)$size[1:857] <- 2
E(i96)$width <- .3
E(i96)$color <- rgb(.5,.5,0,.1)
pdf('i96.2.pdf')
plot(i96, layout=layout.kamada.kawai)
dev.off()
pdf('i96.3.pdf')
plot(i96, layout=layout.fruchterman.reingold.grid)
dev.off()
pdf('i96.4.pdf')
plot(i96, layout=layout.fruchterman.reingold)
dev.off()
g96e <- t(g96) %*% g96 g97e <- t(g97) %*% g97 g98e <- t(g98) %*% g98 i96e <- graph.adjacency(g96e, mode = 'undirected')
E(i96e)$weight <- count.multiple(i96e) i96e <- simplify(i96e)
# Set vertex attributes V(i96e)$label <- V(i96e)$name V(i96e)$label.color <- rgb(0,0,.2,.8) V(i96e)$label.cex <- .6 V(i96e)$size <- 6 V(i96e)$frame.color <- NA V(i96e)$color <- rgb(0,0,1,.5) # Set edge gamma according to edge weight egam <- (log(E(i96e)$weight)+.3)/max(log(E(i96e)$weight)+.3) E(i96e)$color <- rgb(.5,.5,0,egam)
pdf('i96e.pdf')
plot(i96e, main = 'layout.kamada.kawai', layout=layout.kamada.kawai)
plot(i96e, main = 'layout.fruchterman.reingold', layout=layout.fruchterman.reingold)
dev.off()
Group overlap networks and plots
ol96 <- g96e/diag(g96e) ol97 <- g97e/diag(g97e) ol98 <- g98e/diag(g98e)
magall <- ol96 + ol97 + ol98 magall[is.na(magall)] <- 0
magdiag <- apply(cbind(diag(g96e), diag(g97e), diag(g98e)), 1, mean )
magallg <- graph.adjacency(magall, weighted=T) # Degree V(magallg)$degree <- degree(magallg) # Betweenness centrality V(magallg)$btwcnt <- betweenness(magallg)
plot(density(magall))
magallgt1 <- magall magallgt1[magallgt1 < 1] <- 0 magallggt1 <- graph.adjacency(magallgt1, weighted=T) # Removes loops: magallggt1 <- simplify(magallggt1, remove.multiple=FALSE, remove.loops=TRUE)
magallggt1$layout <- layout.fruchterman.reingold(magallggt1) V(magallggt1)$label <- V(magallggt1)$name tkplot(magallggt1)
magallggt1$layout <- tkplot.getcoords(1)
# Set vertex attributes V(magallggt1)$label <- V(magallggt1)$name V(magallggt1)$label.color <- rgb(0,0,.2,.6) V(magallggt1)$size <- 6 V(magallggt1)$frame.color <- NA V(magallggt1)$color <- rgb(0,0,1,.5) # Set edge attributes E(magallggt1)$arrow.size <- .3 # Set edge gamma according to edge weight egam <- (E(magallggt1)$weight+.1)/max(E(magallggt1)$weight+.1) E(magallggt1)$color <- rgb(.5,.5,0,egam)
V(magallggt1)$label.cex <- V(magallggt1)$degree/(max(V(magallggt1)$degree)/2)+ .3 #note, unfortunately one must play with the formula above to get the #ratio just right
pdf('magallggt1customlayout.pdf')
plot(magallggt1)
dev.off()
text_mining.1481501232.txt.gz · Last modified: by hkimscil