summary_of_hypothesis_testing
This is an old revision of the document!
Table of Contents
Hypothesis testing
see also types of error
Basic
see first sampling distribution and z-test
Hypothesis testing, exp
…
- code01
- output01
Loading...
> rm(list=ls())
>
> rnorm2 <- function(n,mean,sd){
+ mean+sd*scale(rnorm(n))
+ }
>
> n.p <- 100000
> m.p <- 100
> sd.p <- 10
> p1 <- rnorm2(n.p, m.p, sd.p)
> m.p1 <- mean(p1)
> sd.p1 <- sd(p1)
>
> p2 <- rnorm2(n.p, m.p+4, sd.p)
> m.p2 <- mean(p2)
> sd.p2 <- sd(p2)
>
> n.s <- 25
> se.z1 <- c(sqrt(var(p1)/n.s))
> se.z2 <- c(sqrt(var(p2)/n.s))
> se.z1
[1] 2
> se.z2
[1] 2
>
> x.p1 <- seq(mean(p1)-5*se.z1,
+ mean(p2)+5*se.z1,
+ length.out = 500)
> x.p2 <- seq(mean(p2)-5*se.z1,
+ mean(p2)+5*se.z1,
+ length.out = 500)
>
> # Calculate the probability
> # density for a normal distribution
> y.p1 <- dnorm(x.p1, mean(p1), se.z1)
> y.p2 <- dnorm(x.p2, mean(p2), se.z2)
>
> # Plot the theoretical PDF
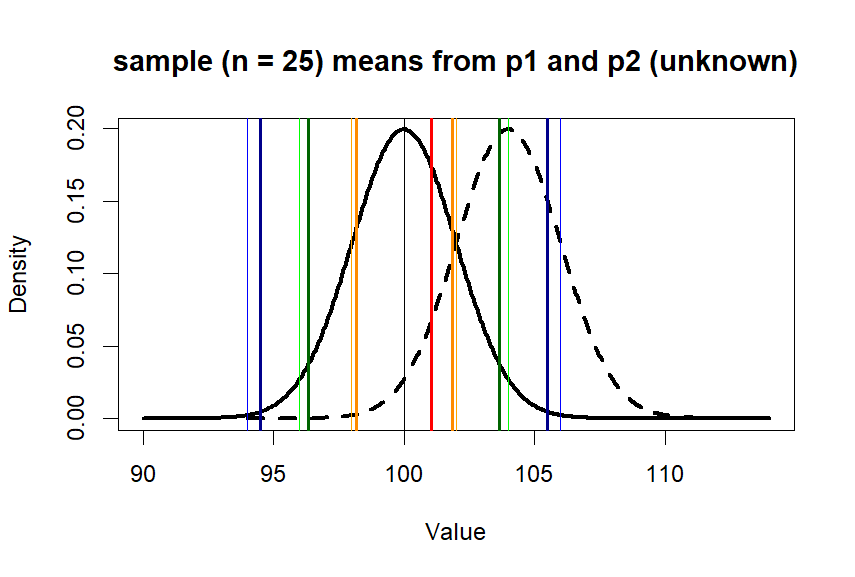
> plot(x.p1, y.p1, type = "l",
+ lwd=3,
+ main = paste0("sample (n = ",n.s, ") means from p1 and p2 (unknown)"),
+ xlab = "Value", ylab = "Density")
> lines(x.p2, y.p2, lty=2, lwd=3)
>
>
> m.p1 <- mean(p1)
> se1 <- c(m.p1-se.z1, m.p1+se.z1)
> se2 <- c(m.p1-2*se.z1, m.p1+2*se.z1)
> se3 <- c(m.p1-3*se.z1, m.p1+3*se.z1)
> abline(v=c(m.p1,se1,se2,se3),
+ col=c('black', 'orange', 'orange',
+ 'green', 'green',
+ 'blue', 'blue'),
+ lwd=1)
>
> treated.s <- sample(p2, n.s)
> m.treated.s <- mean(treated.s)
> abline(v=m.treated.s, col='red', lwd=2)
>
> diff <- m.treated.s-mean(p1)
> diff/se.z1
[1] 0.5253907
> zscore <- abs(diff/se.z1)
> pnorm(zscore, lower.tail = F)*2
[1] 0.5993116
> tscore <- zscore
> pt(tscore, df=n.s-1, lower.tail = F)*2
[1] 0.6041323
>
> # usual way - using sample's variance
> # instead of p1's variance to get
> # standard error value
> se.s <- sqrt(var(treated.s)/n.s)
> se.s
[1] 1.83652
> tscore <- diff/se.s
> tscore
[1] 0.572159
>
>
> se1 <- c(m.p1-se.s, m.p1+se.s)
> se2 <- c(m.p1-2*se.s, m.p1+2*se.s)
> se3 <- c(m.p1-3*se.s, m.p1+3*se.s)
> abline(v=c(se1,se2,se3),
+ col=c('darkorange', 'darkorange',
+ 'darkgreen', 'darkgreen',
+ 'darkblue', 'darkblue'),
+ lwd=2)
>
>
>
>
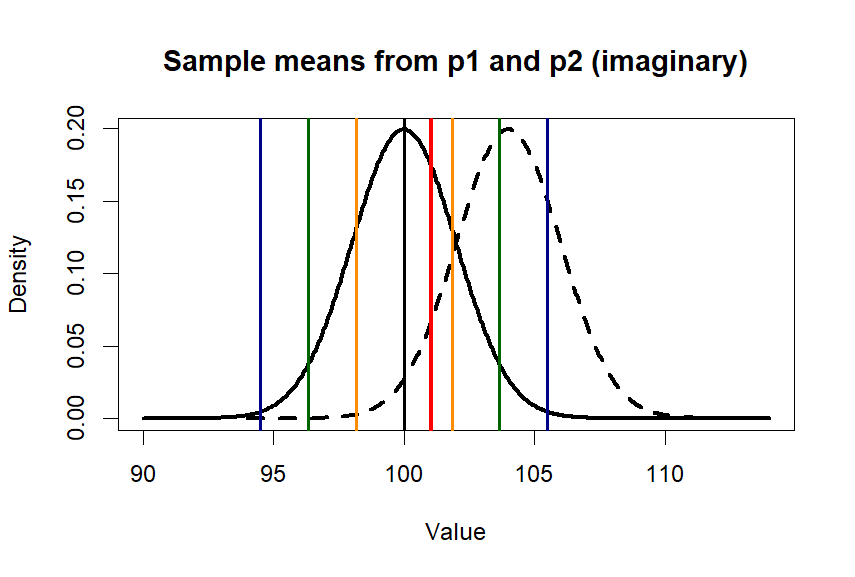
> plot(x.p1, y.p1, type = "l",
+ lwd=3,
+ main = "Sample means from p1 and p2 (imaginary)",
+ xlab = "Value", ylab = "Density")
> lines(x.p2, y.p2, lty=2, lwd=3)
>
>
> m.p1 <- mean(p1)
> se1 <- c(m.p1-se.s, m.p1+se.s)
> se2 <- c(m.p1-2*se.s, m.p1+2*se.s)
> se3 <- c(m.p1-3*se.s, m.p1+3*se.s)
> abline(v=c(m.p1,se1,se2,se3),
+ col=c('black', 'darkorange', 'darkorange',
+ 'darkgreen', 'darkgreen',
+ 'darkblue', 'darkblue'),
+ lwd=2)
> abline(v=m.treated.s, col='red', lwd=3)
> se.s
[1] 1.83652
> se.z1
[1] 2
>
> c(m.treated.s-2*se.s, m.treated.s+2*se.s)
[1] 97.37774 104.72382
> c <- qt(0.975, n.s-1)
> c
[1] 2.063899
> c(m.treated.s-c*se.s, m.treated.s+c*se.s)
[1] 97.26039 104.84117
> m.p2
[1] 104
>
> x.p.est <- seq(m.treated.s-5*se.s,
+ m.treated.s+5*se.z1,
+ length.out = 500)
> y.p.est <- dnorm(x.p.est, m.treated.s, se.s)
>
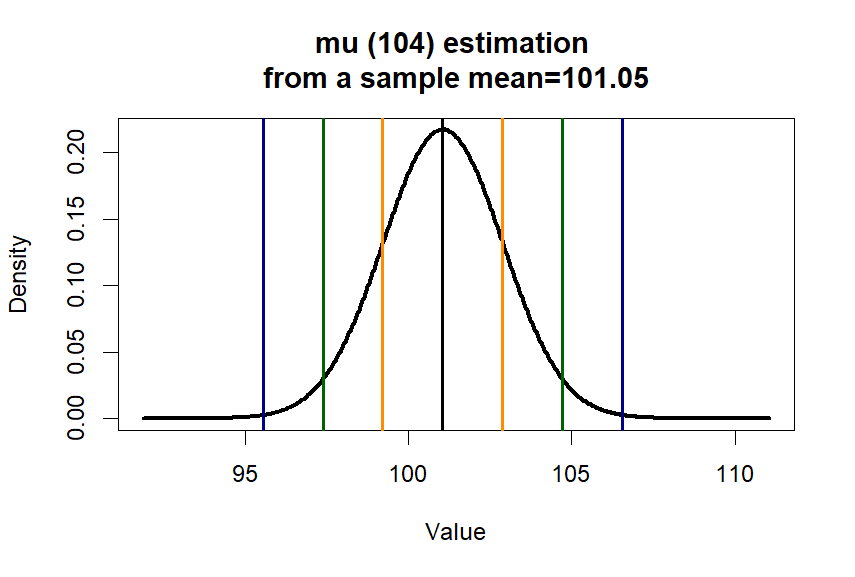
> plot(x.p.est, y.p.est, type = "l",
+ lwd=3,
+ main = paste0("mu (", m.p2, ") estimation \n", "from a sample ", "mean=", round(m.treated.s,2)),
+ xlab = paste0("Value"), ylab = "Density")
> se1 <- c(m.treated.s-se.s, m.treated.s+se.s)
> se2 <- c(m.treated.s-2*se.s, m.treated.s+2*se.s)
> se3 <- c(m.treated.s-3*se.s, m.treated.s+3*se.s)
> abline(v=c(m.treated.s,se1,se2,se3),
+ col=c('black', 'darkorange', 'darkorange',
+ 'darkgreen', 'darkgreen',
+ 'darkblue', 'darkblue'),
+ lwd=2)
>
> tscore <- abs(diff/se.s)
> pt(tscore, df=n.s-1, lower.tail = F) * 2
[1] 0.5725352
> t.test(treated.s, mu=m.p1, var.equal = T)
One Sample t-test
data: treated.s
t = 0.57216, df = 24, p-value = 0.5725
alternative hypothesis: true mean is not equal to 100
95 percent confidence interval:
97.26039 104.84117
sample estimates:
mean of x
101.0508
>
output
>
>
> rm(list=ls())
>
> rnorm2 <- function(n,mean,sd){
+ mean+sd*scale(rnorm(n))
+ }
>
> n.p <- 10000
> m.p <- 100
> sd.p <- 10
> p1 <- rnorm2(n.p, m.p, sd.p)
> m.p1 <- mean(p1)
> sd.p1 <- sd(p1)
>
> p2 <- rnorm2(n.p, m.p+10, sd.p)
> m.p2 <- mean(p2)
> sd.p2 <- sd(p2)
>
> n.s <- 100
> se.z1 <- c(sqrt(var(p1)/n.s))
> se.z2 <- c(sqrt(var(p2)/n.s))
>
> x.p1 <- seq(mean(p1)-5*se.z1,
+ mean(p2)+5*se.z1,
+ length.out = 500)
> x.p2 <- seq(mean(p2)-5*se.z1,
+ mean(p2)+5*se.z1,
+ length.out = 500)
>
> # Calculate the probability
> # density for a normal distribution
> y.p1 <- dnorm(x.p1, mean(p1), se.z1)
> y.p2 <- dnorm(x.p2, mean(p2), se.z2)
>
> # Plot the theoretical PDF
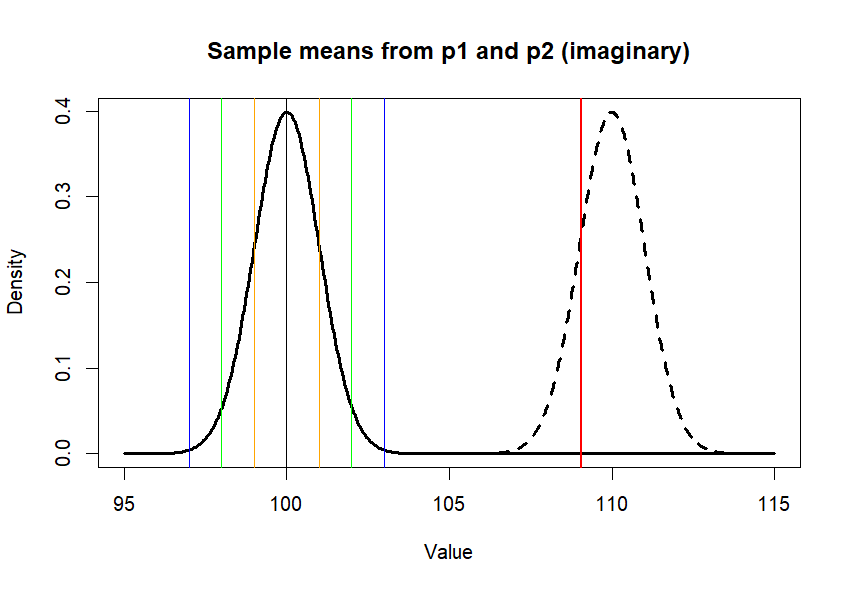
> plot(x.p1, y.p1, type = "l",
+ lwd=3,
+ main = "Sample means from p1 and p2 (imaginary)",
+ xlab = "Value", ylab = "Density")
> lines(x.p2, y.p2, lty=2, lwd=3)
>
>
> m.p1 <- mean(p1)
> se1 <- c(m.p1-se.z1, m.p1+se.z1)
> se2 <- c(m.p1-2*se.z1, m.p1+2*se.z1)
> se3 <- c(m.p1-3*se.z1, m.p1+3*se.z1)
> abline(v=c(m.p1,se1,se2,se3),
+ col=c('black', 'orange', 'orange',
+ 'green', 'green',
+ 'blue', 'blue'),
+ lwd=1)
>
> treated.s <- sample(p2, n.s)
> m.treated.s <- mean(treated.s)
> abline(v=m.treated.s, col='red', lwd=2)
>
> se.z1 [1] 1 > > diff <- m.treated.s-mean(p1) > diff/se.z1 [1] 9.057418 > > # usual way - using sample's variance > # instead of p1's variance to get > # standard error value > se.s <- sqrt(var(treated.s)/n.s) > se.s [1] 1.015243 > diff/se.s [1] 8.921425 > > pt(diff/se.s, df=n.s-1, lower.tail = F) * 2 [1] 2.455388e-14 > t.test(treated.s, mu=m.p1, var.equal = T) One Sample t-test data: treated.s t = 8.9214, df = 99, p-value = 2.455e-14 alternative hypothesis: true mean is not equal to 100 95 percent confidence interval: 107.0430 111.0719 sample estimates: mean of x 109.0574 >
se value and sample size
n.ajstu <- 100000
mean.ajstu <- 100
sd.ajstu <- 10
set.seed(1024)
ajstu <- rnorm2(n.ajstu, mean=mean.ajstu, sd=sd.ajstu)
mean(ajstu)
sd(ajstu)
var(ajstu)
iter <- 10000 # # of sampling
n.4 <- 4
means4 <- rep (NA, iter)
for(i in 1:iter){
means4[i] = mean(sample(ajstu, n.4))
}
n.25 <- 25
means25 <- rep (NA, iter)
for(i in 1:iter){
means25[i] = mean(sample(ajstu, n.25))
}
n.100 <- 100
means100 <- rep (NA, iter)
for(i in 1:iter){
means100[i] = mean(sample(ajstu, n.100))
}
n.400 <- 400
means400 <- rep (NA, iter)
for(i in 1:iter){
means400[i] = mean(sample(ajstu, n.400))
}
n.900 <- 900
means900 <- rep (NA, iter)
for(i in 1:iter){
means900[i] = mean(sample(ajstu, n.900))
}
n.1600 <- 1600
means1600 <- rep (NA, iter)
for(i in 1:iter){
means1600[i] = mean(sample(ajstu, n.1600))
}
n.2500 <- 2500
means2500 <- rep (NA, iter)
for(i in 1:iter){
means2500[i] = mean(sample(ajstu, n.2500))
}
h4 <- hist(means4)
h25 <- hist(means25)
h100 <- hist(means100)
h400 <- hist(means400)
h900 <- hist(means900)
h1600 <- hist(means1600)
h2500 <- hist(means2500)
plot(h4, ylim=c(0,3000), col="red")
plot(h25, add = T, col="blue")
plot(h100, add = T, col="green")
plot(h400, add = T, col="grey")
plot(h900, add = T, col="yellow")
m4 <- mean(means4)
m25 <- mean(means25)
m100 <- mean(means100)
m400 <- mean(means400)
m900 <- mean(means900)
m1600 <- mean(means1600)
m2500 <- mean(means2500)
s4 <- sd(means4)
s25 <- sd(means25)
s100 <- sd(means100)
s400 <- sd(means400)
s900 <- sd(means900)
s1600 <- sd(means1600)
s2500 <- sd(means2500)
sss <- c(4,25,100,400,900,1600,2500) # sss sample sizes
means <- c(m4, m25, m100, m400, m900, m1600, m2500)
sds <- c(s4, s25, s100, s400, s900, s1600, s2500)
temp <- data.frame(sss,
means,
sds)
temp
ses <- rep (NA, length(sss)) # std error memory
for(i in 1:length(sss)){
ses[i] = sqrt(var(ajstu)/sss[i]) # std errors by theorem
}
data.frame(ses)
se.1 <- ses
se.2 <- 2 * ses
lower.s2 <- mean(ajstu)-se.2
upper.s2 <- mean(ajstu)+se.2
data.frame(cbind(sss, ses, lower.s2, upper.s2))
# 12/2 lecture
# note that we draw the statistical calculation
# by "diff/se" = "diff/random_error"
n <- 80
mean.sample <- 103
sample <- rnorm2(n, mean.sample, sd.ajstu)
mean(sample)
sd(sample)
diff <- mean.sample - mean.ajstu # this is actual difference
se <- sd.ajstu / sqrt(n) # this is random error
t.cal <- diff/se
t.cal
qnorm(0.025, lower.tail = F)
qnorm(0.01/2, lower.tail = F)
qt(0.05/2, n-1, lower.tail=F)
t.test(sample, mu=mean.ajstu)
# or we obtain the exact p value
p.value <- pt(t.cal, n-1, lower.tail = F)
p.value*2
summary_of_hypothesis_testing.1764517433.txt.gz · Last modified: by hkimscil